Gene Variants Overview
Overview
Genetic variants are central to understanding the causes and mechanisms of rare diseases. The Gene Variants Overview brings together data on reported sequence changes, linking them to genes, diseases, and clinical features. By consolidating these sources, the feature supports exploration of variant frequency, functional impact, and disease associations. Use the tabs below to explore data from public databases, manually curated literature, AI-mined literature, and protein visualization.
*The information is provided to advance research and is not intended for clinical use.
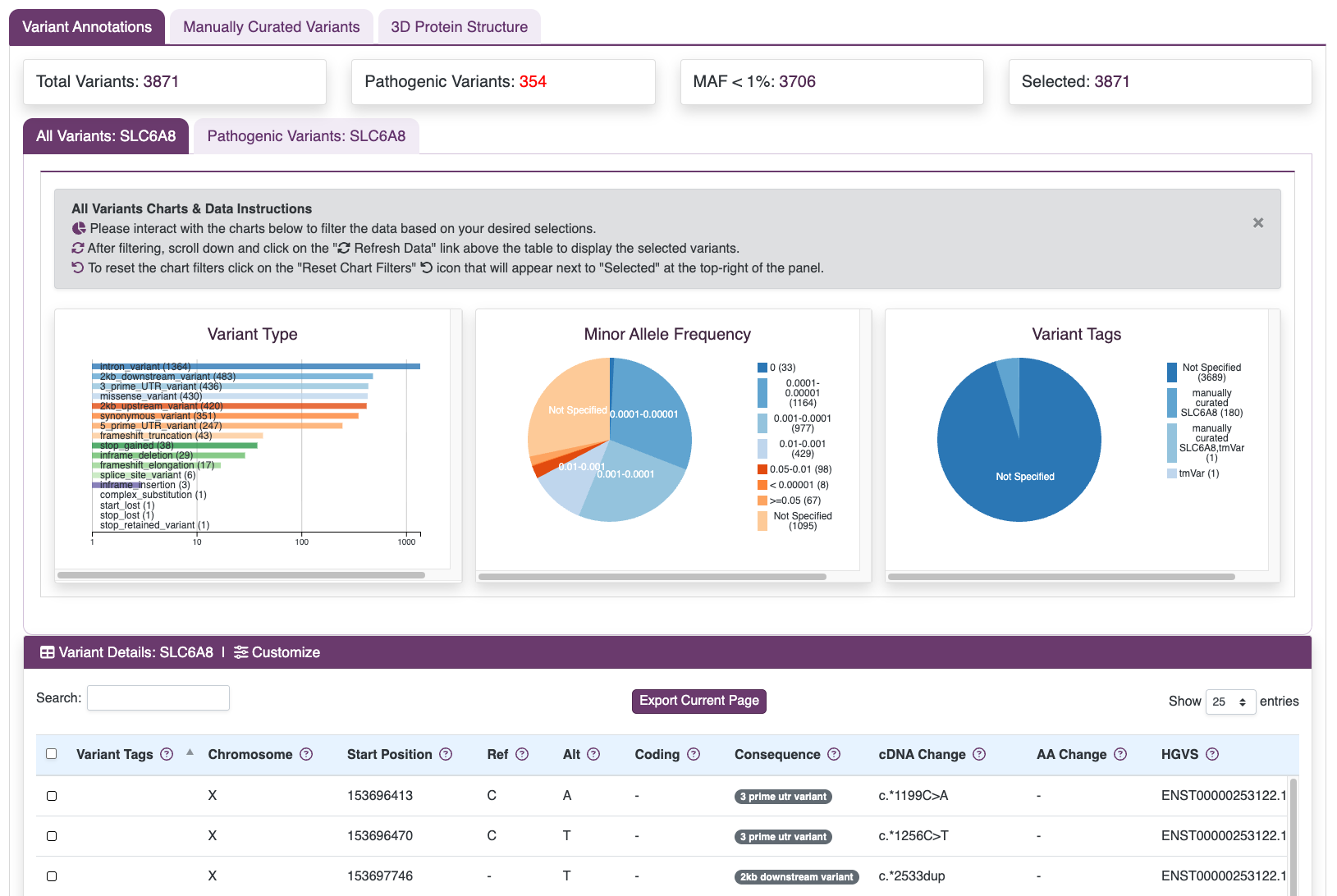
Variants from widely used genomic resources, including ClinVar, dbSNP, and gnomAD are downloaded and integrated. Annotations are added to provide classifications, allele frequencies, and supporting references, offering critical context for each variant.
Select or Search for a Gene Symbol:
View Public Variants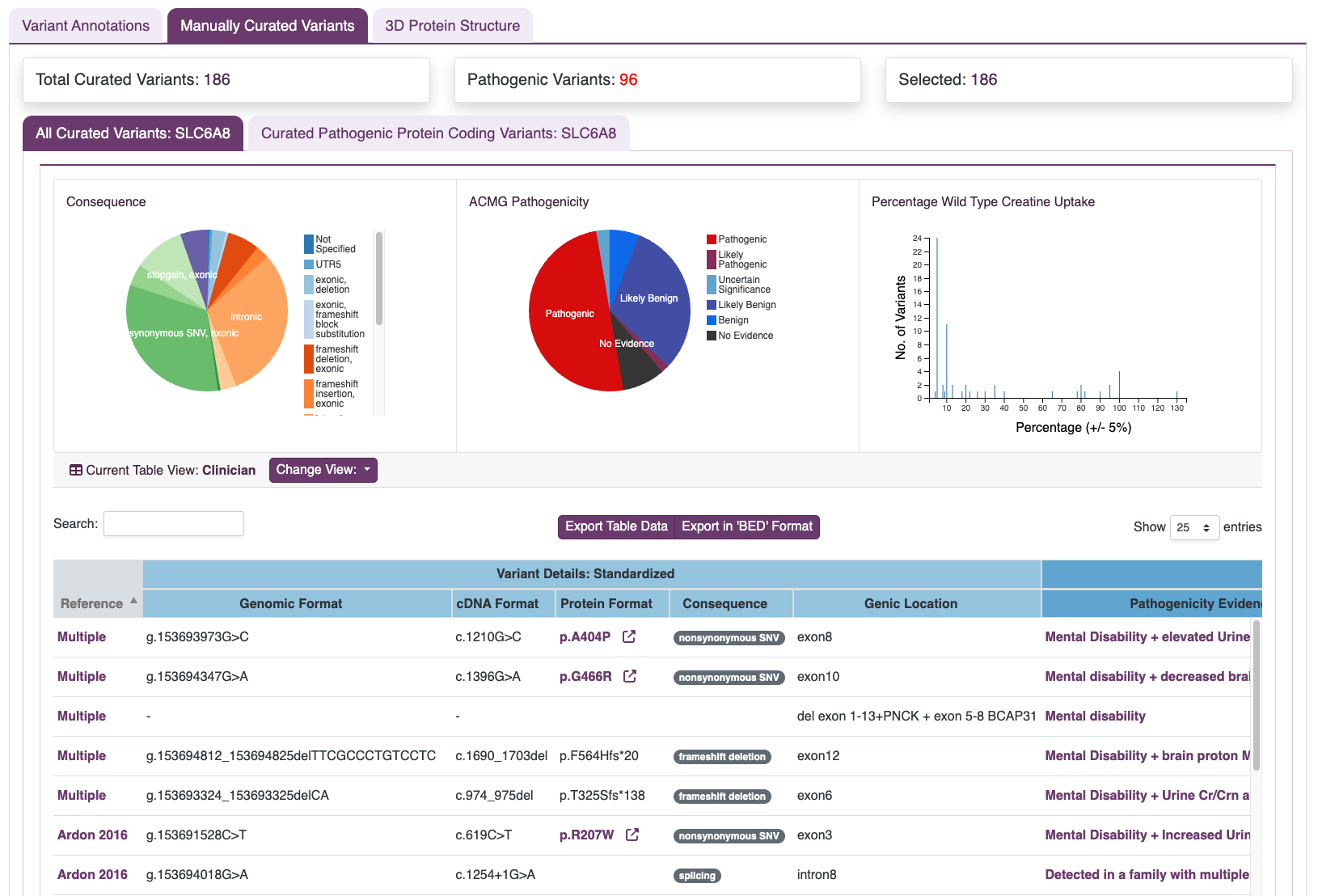
Rare diseases may individually be rare but are collectively common. Information on pathogenic variants is vital for disease diagnosis as well as research on therapeutic approaches. Despite being published in the literature, pathogenicity data on patient variants may not be accessible through a public resource. Our researchers have read peer reviewed manuscripts to create a curated list of published variants. The curated details are reported as a highly annotated dataset of variants with clinical context and functional details including assessments and reported biological effects. Currently, curated variants are available for SLC6A8 (X-linked Creatine Transporter Deficiency), with additional curation in progress for Farber Disease and related genes.
SLC6A8 (X-linked Creatine Transporter Deficiency): manually curated variants with clinical details, inheritance, pathogenicity calls, and functional readouts (e.g., creatine uptake).
View Curated SLC6A8 VariantsTo complement expert review, RARe-SOURCE® employs automated text-mining and machine learning to identify additional variant mentions across the biomedical literature. This approach highlights candidate variants, phenotype associations, and supporting references that may not yet appear in curated datasets. All mined results remain linked to their source publications, expanding discovery potential while preserving transparency and traceability.
Select or Search for a Gene Symbol:
Submit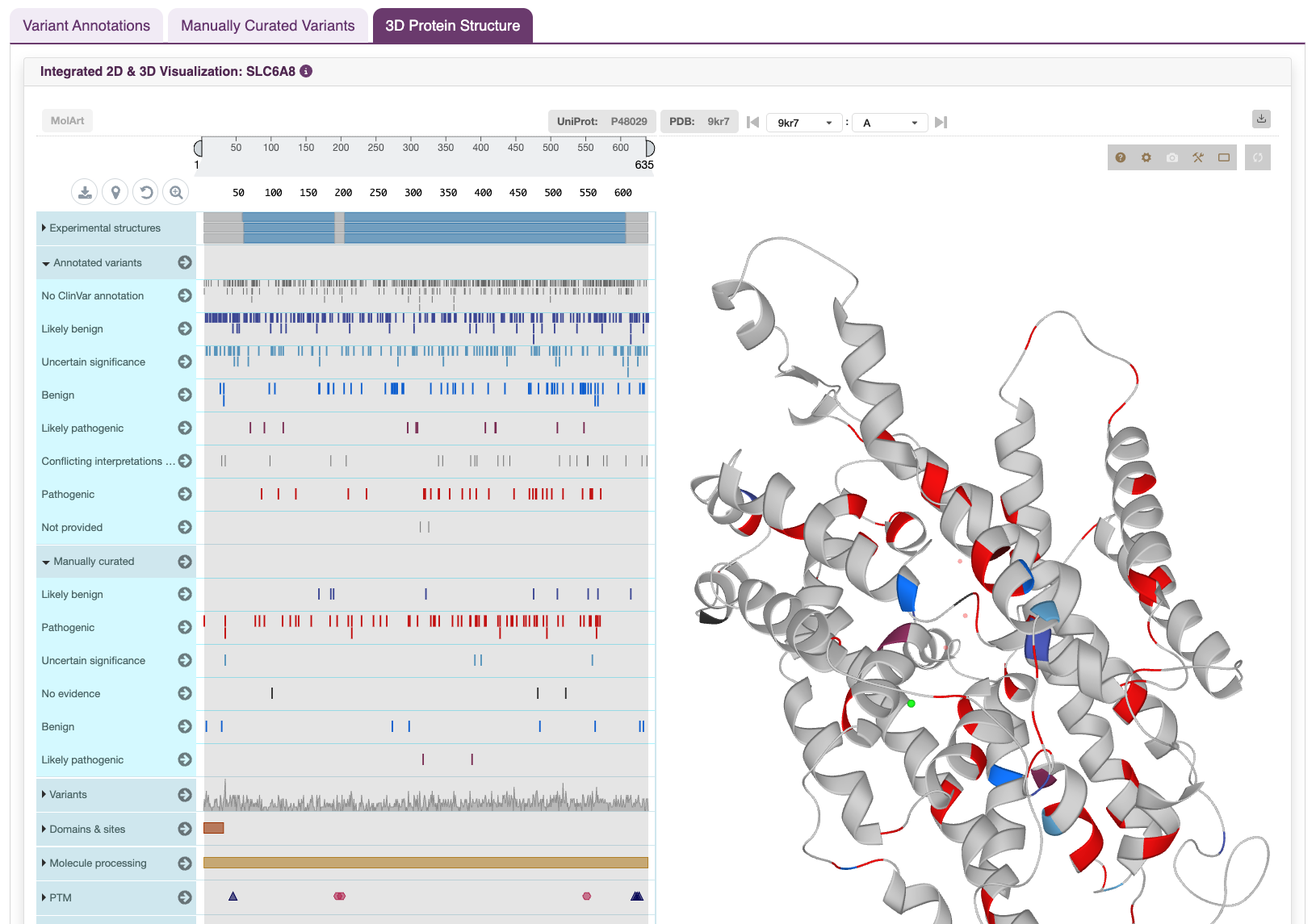
The Protein Visualization feature provides a quick view of domain architecture, key residues, and reported variant locations for a selected gene’s canonical protein. Annotated variants from public databases and manual curation are integrated for visual insights.
